Spatial validation, variable selection and parameter tuning in one go
This page serves as a short introduction to spatialMaxent before we will start doing the modeling. There are a few things that have changed compared to maxent and we will explain here what you should look out for.
The data structure
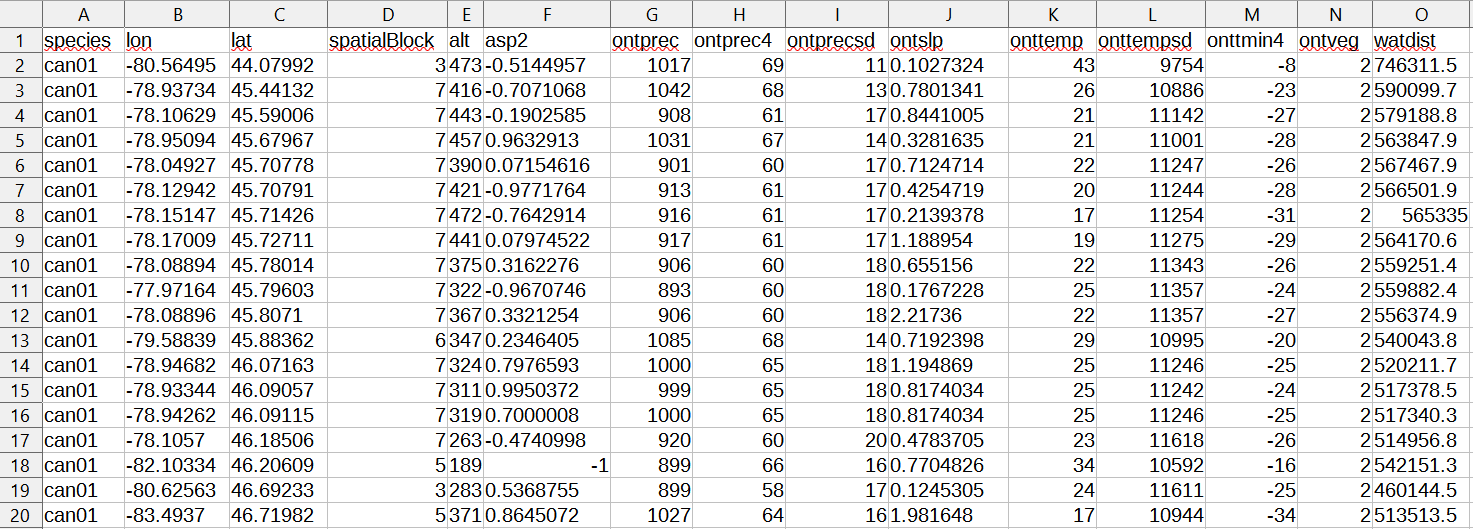
First of all the data structure changes slightly insofar as it is necessary to provide the samples file with a fourth column containing the association of each point with a spatial fold as integer value. If the SWD data format is chosen also the environmental layers .csv file needs to have an additional column for the folds, but no values are required here. Note that in spatialMaxent contrary to Maxent the processing of several species at once from the same samples file is not supported.
The csv file we created on the previous pages are already in the right format. The first three columns contain the species, longitude and latitude and the fourth the folds created with the blockCV package. After these mandatory columns all columns with environmental information follow. This should look like this:
Spatial Validation
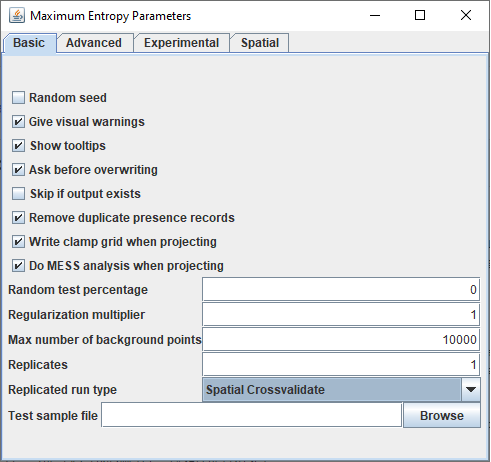
The spatial validation is a new option that can be chosen in the basic tab as Replicated run type. If the spatial crossvalidate option is used the setting of replicates will have no effect anymore as the number of replicates will be set to the number of distinct folds in the sample data.
Tuning options
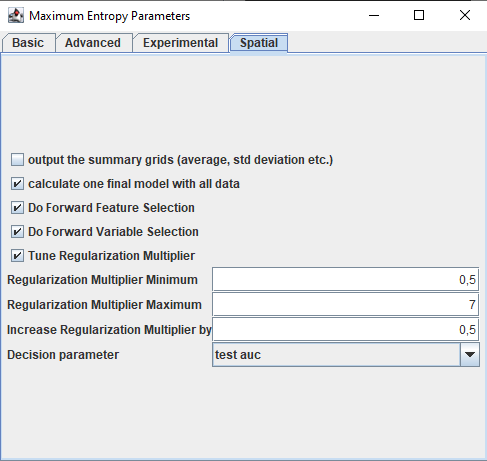
The nice thing about spatialMaxent is that all tuning tasks are available at once with just a few clicks. In addition to the selection of the replication multiplier and the features, a variable selection is also carried out. The following functionalities are available in spatialMaxent: Forward-Variable-Selection, Forward-Feature-Selection and regularization multiplier tuning. All the tuning procedures can be found in the GUI in a new tab of the settings called spatial. Each of the steps variable selection, feature selection and regularization multiplier tuning can be omitted but we highly recommend to do all of them.
Forward Variable Selection (FVS)
The Forward Variable Selection as designed by Meyer et al. (2018) is a variable selection method particularly designed for spatial data. FFS trains all possible 2 variable combinations, choses the best one and then trains these two variables together with each of the remaining variables and repeats this step until the models show no increase in performance. FVS will be run in parallel if threads>1
Forward Feature Selection (FFS)
The Forward Feature Selection follows the same structure as the FVS. It trains one model with each feature (hinge, linear, threshold, product, quadratic). Then the best one is chosen and the other features are tested in combination with it until no increase in model performance is observed. Note that the setting of features in the main maxent tab (Auto features, Hinge features, etc.) will be ignored by spatialMaxent if an FFS is done as all features are tested one after another.
Regularization multiplier tuning
The regularization multiplier tuning has three input parameters: The lowest regularization multiplier to be tuned the highest regularization multiplier to be tuned and the steps in which the regularization multiplier is increased from the minimum regularization multiplier to the highest regularization multiplier. e.g.(RMMin=1,RMMax=5 and RMIncrease=0.5 will try the following regularization multipliers: 1 1.5 2 2.5 3 3.5 4 4.5 5).
Which model is the best one can be either determined based on the test gain or the test auc value (decision parameter).
Optional output of summarized grids
With the setting cvGrids it is possible to omit the output of the summarized html page and grids of cross-validation or spatial cross-validation. The grids average, max, median, min and stddev will not be written to disc if this parameter is set to false.
Calculate a final Model
The setting finalModel can be used to train one model at the end with the selected variables, the selected features and the selected regularization-multiplier and all presence records.
general notes
The FFS, FVS and regularization multiplier tuning all have large processing times. Especially if the FVS is done and there are lots of input variables alone in the first step of the FVS this can lead to a large amount of models to be trained if the input consists of 10 variables 45 models are trained in the first step of the FVS if it consists of 20 variables it increases to 190 models. Just keep in mind, that the whole process can take a while. In this case consider trying out the parallel processing of the FVS by setting threads larger than 1.
If you are working on the command line these are the parameters to use:
| Name | Type | Default |
|---|---|---|
| fvs | boolean | true |
| ffs | boolean | true |
| tuneRM | boolean | true |
| RMMin | double | 0.5 |
| RMMax | double | 7 |
| RMIncrease | double | 0.5 |
| cvGrids | boolean | false |
| finalModel | boolean | true |
