Spatial validation
spatialMaxent performs a spatial cross-validation during the model training. When working with environmental data a spatial validation strategy hast to be chosen that accounts for spatial autocorrelation in the data (Ploton et al. 2020). If the data is partitioned random the models can be easily overfitted on the training data which can lead to very complex models that generalize badly. In addition to the internal validation strategy, the test data not included in the model must also be spatially independent, which is why we decided not to validate the models with the PA data here. To create spatial independent folds for spatial cross-validation we will now use the blockCV package by Valavi et al. 2018 to part the data into seven folds.
Of the seven presence records folds, five are used to train the models and two are used as independent test data. Since one of the most crucial points in spatial validation is the distribution of test and training points, the selection of the folds that are removed from the data for testing can lead to large differences in the determination of the model quality. If the test data is randomly drawn from all available folds, the metrics calculated for the models are only of limited value. To get a comprehensive picture of which method performs best on the dataset, we propose to remove this effect from the results by using a Forward Fold Metric Estimation (FFME). For this purpose, models are calculated for all possible combinations of training and test data and the models are evaluated with the respective independent test data. The median of all results metrics is then used to assess the quality of the method (blue box in the Figure below). Each of the PO points will be part of the test and training data at some point while at the same time the models can be evaluated with independent data.
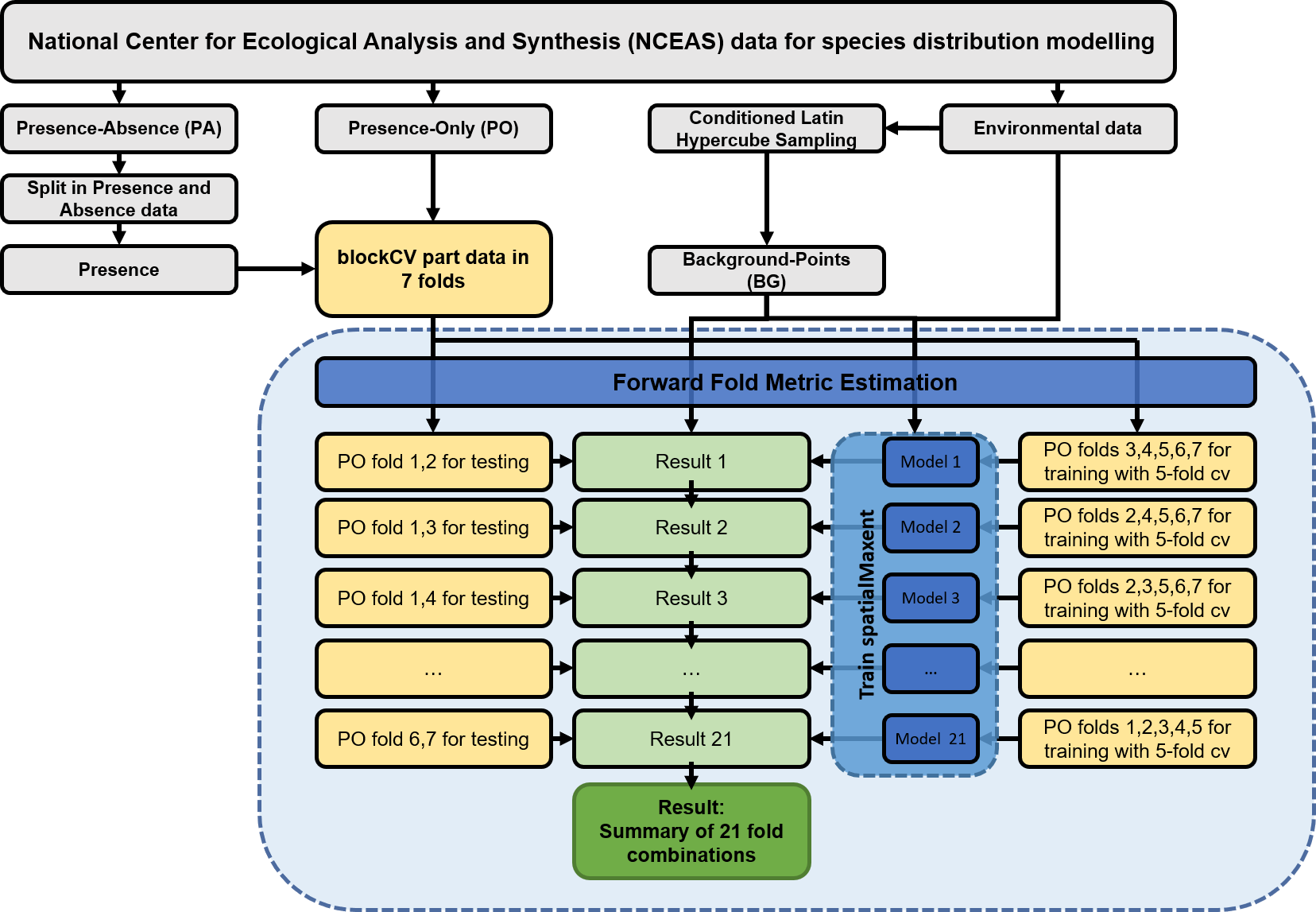
In the next script we will first create spatial blocks and then create training and test files for each step of the FFME and save them separately.
The spatial blocks generated with this code for the species can01 can be seen in the map displayed at the bottom of the page.
